Goal
To establish a microflow-nanospray ESI-MS (MnESI-MS) platform for liquid chro- matography-mass spectrometry (LC-MS) analysis of oligonucleotides, that achieves high sensitivity and high robustness, using Newomics® MnESI sources, M3 multinozzle emitters, and flow splitting kits (as needed for high-flow HPLC/UPLC systems).
Introduction
Oligonucleotide therapeutics including single-stranded antisense oligonucleotides (ASOs) and double-stranded siRNAs are rapidly growing. Compared to traditional hybridization-based methods such as ELISA and PCR, mass spectrometry (MS) analysis of oligonucleotide is more specific because it can discriminate and quantify not only the full-length products but also their impurities and metabolites. High-flow ion-pairing reversed-phase chromatography has become the LC method of choice for oligonucleotide analysis [1]. However, current LC-MS methods have the challenges of low sensitivity for negative ionization mode, and high contamination to the MS because of the ion-pairing reagents required for LC separation of oligos. The Newomics award-winning silicon multinozzle emitters (M3 emitter) split the incoming microflow eluent into multiple nanoflows at each nozzle, thereby significantly enhancing the ionization efficiency and reducing the matrix effects for ESI-MS ([2-5] and Newomics Application Notes [6]). In this Application Note, we demonstrate a new microflow-nanospray ESI-MS platform for LC-MS analysis of a 21 mer DNA oligonucleotide. Compared to the high-flow LC-MS, we obtained over 30-fold improvement in sensitivity, with a CV of less than 5%. Our MnESI-MS platform is amenable for a microflow LC system, as well as a high-flow LC system when interfaced with a Newomics flow splitting kit. Our platform for oligonucleotide analysis increases the sensitivity through increased ionization efficiency at the low flow rate, as well as reduces the MS contamination because of fewer ion-pairing reagents entering the mass spectrometers.
Methods
1. Reagent and sample preparation
The 21 mer DNA oligo CAG TCG ATT GTA CTG TAC TTA was synthesized and desalted at Integrated DNA Technologies. Ethylenediaminetetraacetic acid solution (EDTA, Cat. # 03690-100ML), HPLC-grade water and methanol were purchased from Sigma-Aldrich (St. Louis, MO). LC-MS grade Triethylamine (TEA) was purchased from RICCA chemical company (Cat. # RMB11205), and 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP) was purchased from Honeywell Fluka (Cat. # 42060-50ML). DNA oligo was diluted in 1mM EDTA at 10 ng/μl before LC-MS analysis.
2. MnESI-MS platform for LC-MS analysis
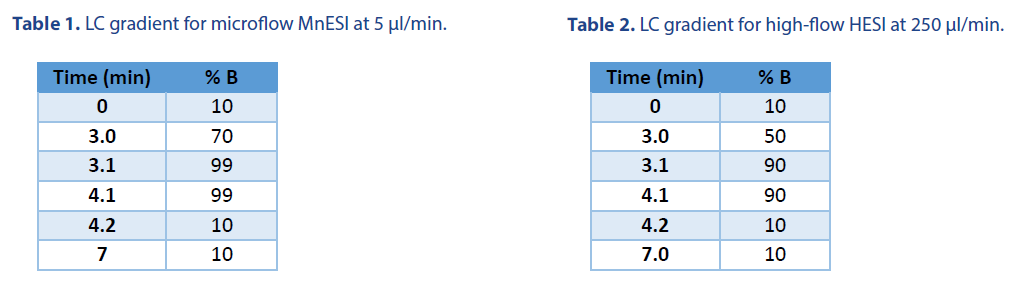
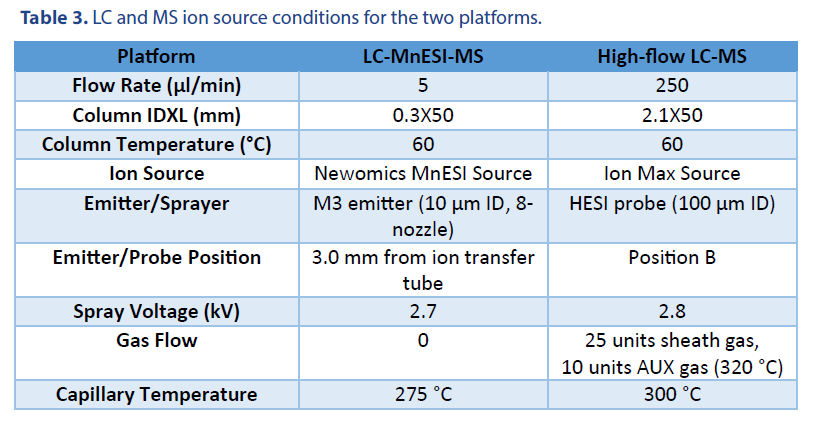
10 ng of 21 mer oligo DNA was injected using a 1 μl full loop injection method. The microflow LC was performed with a BEH C18 column (Waters Cat. # 186009257). The high-flow LC was performed with an XBridge Oligonucleotide Premier C18 column (Waters Cat. # 186009836). Both columns have the same 130 Å pore size of C18 beads. The LC gradients were listed in Table 1 and Table 2, respectively. Mobile phase A was 1.6 mM TEA and 20 mM HFIP, and Mobile phase B was 1.6 mM TEA and 20 mM HFIP in 50% methanol.
Mass spectra were acquired on a Thermo Fisher Orbitrap Q Exactive Plus mass spectrometer interfaced with an UltiMate 3000 RSLCnano UPLC system (Thermo Fisher Scientific). MnESI ion source was either directly connected to the microflow LC column or to a high-flow column through a Newomics® flow splitter.
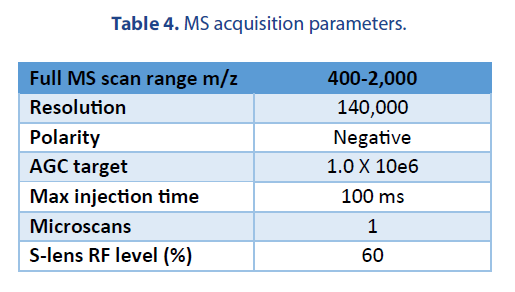
We performed a comparison of the LC-MS analysis among two platforms. The LC and MS source conditions were listed in Table 3.
The following emitters were used in our studies: a Thermo Fisher high-flow needle insert assembly (HESI high-flow, single nozzle, 100 µm ID, catalog # Opton 53010), and a Newomics® M3 emitter (8 nozzles, 10 µm ID, catalog # E8N10MU01).
The MS acquisition parameters for the two platforms were the same and listed in Table 4.
3. Data Analysis
Data were analyzed using Xcalibur software and Biopharma Finder 2.0 software (Thermo Fisher Scientific). Charge deconvolution was processed by Biopharma Finder 2.0 with the Xtract function.
Results and Discussion
1. Optimization of microflow LC-MS for oligonucleotide analysis
We used a Waters microflow C18 column in our oligonucleotide analysis. We found that the concentrations of TEA and HFIP are important to achieve a good chromatography peak for the microflow column. The LC mobile phases with the ion-pairing reagent need to be freshly prepared and typically used within a day for direct sensitivity comparison. Figure 1A shows the representative extracted ion chromatogram of the most abundant charge state (10-) from each platform. We observed a slight charge envelope shift towards lower m/z (higher charge states) for MnESI compared to high-flow HESI (Figure 1B). In addition, the majority of the MS signals from MnESI were from the three higher charge states (9- to 11-), suggesting better and more homogenous ionization of oligo DNA for microflow LC-MS. By either preconditioning the microflow column with EDTA or preparing an oligonucleotide sample in 1 mM TDTA, we found the salt adducts (Na+, K+) were almost completely removed, confirming the MnESI workflow suitable for accurate quantification of oligonucleotides.
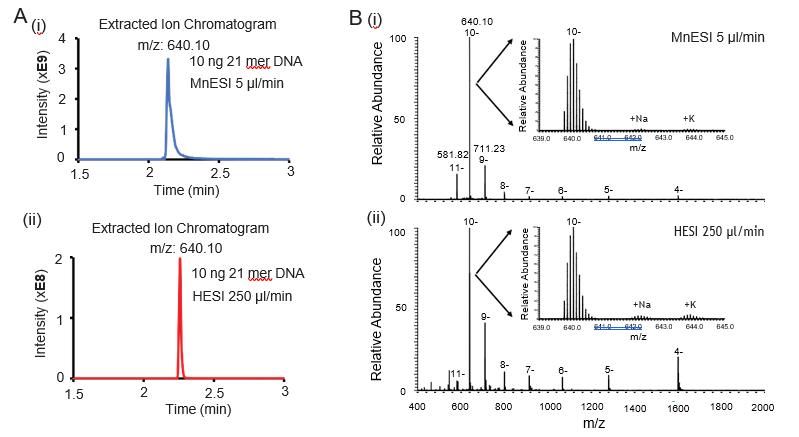
400-2000 for MnESI (i) and high-flow HESI (ii). Compared to high-flow HESI, MnESI shows a charge envelope that is concentrated towards the higher m/z region (9- to 11-). The zoom-in views show the isotopic envelope of the 10- charge state and low salt adducts.
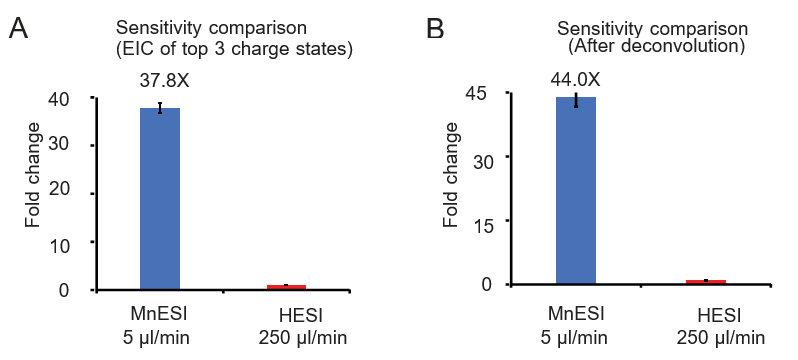
2. MnESI-MS platform significantly improves sensitivity for oligonucleotide analysis
We examined the sensitivity gain of MnESI at 5 µl/min over the conventional HESI platform at 250 µl/min using 10 ng on-column DNA oligo under the optimized conditions for each analysis.
Two quantification methods were used for comparison including the peak area of the extracted ion chromatogram of the top 3 charge states (9- to 11-) and using the charge deconvolution method (4- to 11-). We achieved significant and consistent sensitivity gain from MnESI over high-flow HESI, specifically an average of 37.8-fold using the EIC peak area (Figure 2A) and 44-fold using the charge deconvolution (Figure 2B). In addition, we found very good reproducibility from both platforms with their CVs of less than 5%.
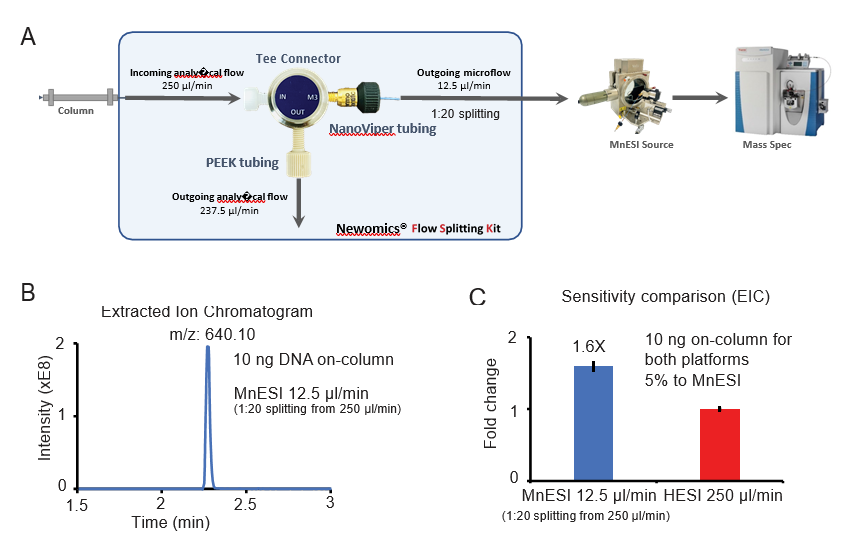
3. MnESI-MS platform seamlessly integrates with the high-flow LC system for oligonucleotide analysis
Since the traditional workflow for oligonucleotide analysis is high-flow LC-MS by HESI, we evaluated the performance of coupling MnESI with a high-flow LC system and 2.1 mm ID column using a Newomics® post-column splitting kit (Figure 3A). A T-splitter was used to split the analytical flow of 250 µl/min from the LC column down to a microflow of 12.5 µ l/min (1:20 splitting) and delivered to a 10 µm-ID, 8-nozzle M3 emitter for spray into MS. A representative extracted ion chromatogram of the most abundant charge state (10-) is shown in Figure 3B. Compared to the conventional high-flow method without the post-column splitting, this new method by flow splitting to M3 emitters achieved slightly better sensitivity (~1.6 fold), even though only 5% of oligonucleotide was delivered to MS (Figure 3C). Our MnESI-MS workflow with high-flow post-column splitting allows the application of MnESI for microflow without the need to change the high-flow LC system. In addition, the majority of the ion-pairing reagent from the high-flow column was diverted to waste rather than going into the mass spectrometer, minimizing the contamination of the ion cone and C-trap of the QE instrument. This will significantly improve the assay robustness and instrument stability for oligonucleotide quantitation.
Conclusion
We have established a new MnESI-MS platform for sensitive LC-MS analysis of oligonucleotides. By interfacing with an Orbitrap Q Exactive Plus mass spectrometer, Newomics® MnESI platform has achieved high sensitivity for analysis of oligonucleotides. We demonstrate the following significant advantages of our LC-MnESI-MS platform using M3 emitters over the traditional high-flow LC-MS platform using HESI:
- More than 30-fold sensitivity gain using the microflow LC-MS at the 5 ul/min flow rate.
- Sensitivity increased 1.6-fold even after 1:20 flow splitting of the high flow, with only 5% material entering the MS.
- Flow splitting reduced contamination due to much less ion-pairing reagent entering the MS.
Reference
- Sutton, M. et al. (2020) Current State of Oligonucleotide Characterization Using Liquid Chromatography-Mass Spectrometry: Insight into Critical Issues. J Am Soc Mass Spectrom 31 (9), 1775-1782.
- Kim, et al. (2007) Microfabricated monolithic multinozzle emitters for nanoelectrospray mass spectrometry. Anal Chem 79 (10), 3703-7.
- Mao, P. et al. (2011) Multinozzle emitter arrays for nanoelectrospray mass spectrometry. Anal Chem 83 (15), 6082-9.
- Mao, P. et al. (2013) Multinozzle emitter array chips for small-volume proteomics. Anal Chem 85 (2), 816-9.
- Chen, et al. (2018) Quantitation of Intact Proteins in Human Plasma Using Top-Down Parallel Reaction Monitor- ing-MS. Anal Chem 90 (18), 10650-10653.